|
Bacterial Interactomes: Interacting Protein Partners Share Similar Function and Are Validated in Independent Assays More Frequently Than Previously Reported
Maxim Shatsky, Simon Allen, Barbara L. Gold, Nancy L. Liu, Thomas R. Juba, Sonia A. Reveco, Dwayne A. Elias, Ramadevi Prathapam, Jennifer He, Wenhong Yang, Evelin D. Szakal, Haichuan Liu, Mary E. Singer, Jil T. Geller, Bonita R. Lam, Avneesh Saini, Valentine V. Trotter, Steven C. Hall, Susan J. Fisher, Steven E. Brenner, Swapnil R. Chhabra, Terry C. Hazen, Judy D. Wall, H. Ewa Witkowska, Mark D. Biggin, John-Marc Chandonia, and Gareth Butland
Molecular & Cellular Proteomics (2016)15:1539-1555
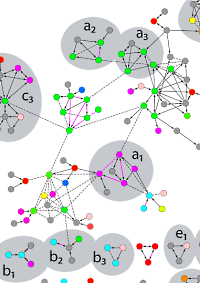
The authors conducted an affinity purification-MS survey of the sulfate-reducing bacterium D. vulgaris and also reanalyzed nine yeast-2-hybrid and AP-MS screens for other bacteria. By using more stringent criteria for assessing the quality of protein interactomes, they concluded that the number of bona fide interactions from the earlier screens is limited to hundreds and not the thousands claimed.
The majority of AP samples were analyzed by parallel gelfree and gel-based workflows. In the gel-free approach, AP isolated proteins were digested with trypsin utilizing a 96-well PVDF membrane-based protocol and analyzed by LC MS/MS. Combined with their data analysis strategy they identified 459 high confidence protein-protein interactions for D. vulgaris and 391 from an existing AP-MS dataset for E. coli, many of which are supported by low throughput data from the literature. |
 |