|
Approach for identifying HLA-DR bound peptides from scarce clinical samples
Tina Heyder, Maxie Kohler, Nataliya K. Tarasova, Sabrina Haag, Dorothea Rutishauser, Natalia V Rivera, Charlotta Sandin, Sohel Mia, Vivianne Malmstrom, Asa M. Wheelock, Jan Wahlstrom, Rikard Holmdahl, Anders Eklund, Roman A. Zubarev, Johan Grunewald and Anders Jimmy Ytterberg
Molecular & Cellular Proteomics (2016) published online July 24, 2016
In the field of immunopeptidomics, large numbers of cells are often required due to the complexity of the samples and the limitations of the methods. Here the authors have developed an improved method for the identification of HLA-DR bound peptides from low cell numbers.
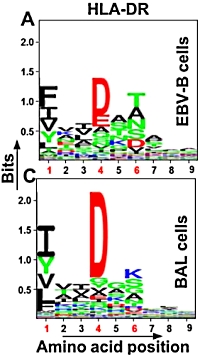
The authors used cell populations consisting of Epstein-Barr-immortalized B cells and broncoalveolar lavage cells obtained from patients with sarcoidosis. The optimized approach included cell lysis, immunoprecipitation of HLA-DR, elution of HLA-DR bound peptides, desalting and peptide concentration, and identification by mass spectrometry followed by peptide filtering.
This resulted in the identification of 507 peptides (from 75 proteins) in the EBV-B cells and 1,434 peptides (from 215 proteins) in the BAL samples with an average yield of 10.1 peptides/million cells. This yield represents a 1 to 2 order of magnitude improvement over previous studies. |
 |