|
PCoM-DB Update: A Protein Co-Migration Database for Photosynthetic Organisms
Atsushi Takabayashi, Saeka Takabayashi, Kaori Takahashi, Mai Watanabe, Hiroko Uchida, Akio Murakami, Tomomichi Fujita, Masahiko Ikeuchi and Ayumi Tanaka
Plant Cell Physiol. (2016) online: December 22, 2016
To facilitate the understating of protein complexes in cellular processes, the authors have developed a searchable database of protein co-migration for photosynthetic organisms.
They used blue-native PAGE for separation, cut the gel strips into ~60 pieces, subjected them to in-gel digestion, and analyzed by LC-MS/MS with protein identification by Mascot. Protein abundances were estimated by spectral counting, using the exponentially modified protein abundance index (emPAI),
as implemented in Mascot.
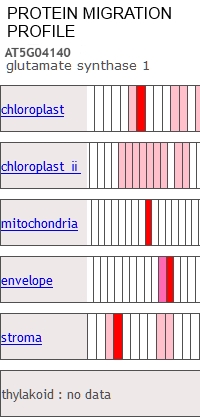
The Protein Co-Migration Database (PCoM-DB) provides prediction tools for protein complexes. PCoM-DB displays migration profiles for any given protein of interest, and allows users to compare them with migration profiles of other proteins, showing the oligomeric states of proteins and, thus, identifying potential interaction partners.
This paper covers the addition of data to the PCoM-DB from bryophytes, green algae, cyanobacteria, as well as Arabidopsis organelles, including intact chloroplasts, chloroplast stroma, the chloroplast envelope, and mitochondria.
|
 |