|
Selected reaction monitoring approach for validating peptide biomarkers
Qing Wang, Ming Zhang, Tyler Tomita, Joshua T. Vogelstein, Shibin Zhou, Nickolas Papadopoulos, Kenneth W. Kinzler, and Bert Vogelstein
PNAS 2017 114 (51) 13519-13524; published online December 4, 2017
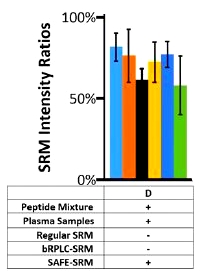
The authors have developed a selected reaction monitoring (SRM)-based method for the discovery and validation of peptide biomarkers for ovarian cancer. It utilizes multiple steps of fractionation and separation as well as synthesis of target peptides, and they call it "sequential analysis of fractionated eluates" - SAFE-SRM.
The study was executed in three phases
- Phase 1, global proteomic profiling of plasma samples from cancer patients and healthy individuals, yielding 641 candidate peptide markers from 188 genes
- Phase 2, implementation of a SRM-based assay to evaluate each of the 641 candidate peptide markers in additional plasma samples, yielding two peptides from peptidyl-prolyl
cis-trans isomerase A as promising biomarkers
- Phase 3, evaluation of the performance of these two peptides in an independent set of cancer patients and controls using SAFE-SRM.
To provide validation a separate cohort of 73 cases, consisting of plasma from 35 ovarian cancer cases and 38 samples from healthy individuals or patients with other cancer types, was tested against the two PIPA peptides. Twenty (57.1%; 95% CI, 40-73%) of the 35 plasma samples from ovarian cancer cases scored positive for one or both of these peptides. None of the 14 samples from normal individuals scored positive.
|
 |